Molecular Mechanisms of Movement Disorders

Team: Aleksandar Rakovic (Group leader), Britta Maier (MTA), Arian Hach (PhD student), and Kerstin Tanzer (MD student)
The main focus of the team “Molecular Mechanisms of Movement Disorders” is the characterization of molecular pathways involved in the pathophysiology of Parkinson’s disease (PD)and Dystonia using human cellular models (human dermal fibroblasts, neuroblastoma cell lines, and induced pluripotent stem cell (iPSC)-derived neurons). We investigate the interaction between the PD-associated genes mitochondrial kinase PINK1 (PTEN-induced putative kinase 1) and the ubiquitin E3 ligase Parkin and their role in the removal of damaged mitochondria by autophagy (mitophagy). Previoiusly we discovered that PD-causing mutations in PINK1 impair recruitment of Parkin to damaged mitochondria and demonstrated the role of Parkin in the ubiquitination of outer mitochondrial membrane proteins as an initial step in mitophagy. Currently, we focus on depicting the exact role of the ubiquitin proteasome system and the lysosomal system in mitophagy.
Dr. Rakovic is a head a platform for genome editing using CRISPR/Cas9 system in various cellular models and Zebrafish. For this, we are using episomal vectors for expressing both Cas9 protein and guide RNA (gRNAs) as well as in-vitro synthesized “capped” Cas9 mRNA and gRNAs for an integration-free approach. We routinely generate isogenic knock-in and reporter iPSC lines and knockout Zebrafish (Danio rerio) models of using CRISPR/Cas9.
Projects are funded by intramural grant support, the DFG, and the Collaborative Center for XDP (CCXDP).
We collaborate with several national and international colleagues including Prof. Dimitri Krainc (Northwestern University, Feinberg School of Medicine, USA), Prof. Reinhard Köster (Technical University Braunschweig), the SysMed Consortium (Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Luxembourg), Prof. Frank Kaiser (University Hospital Essen).
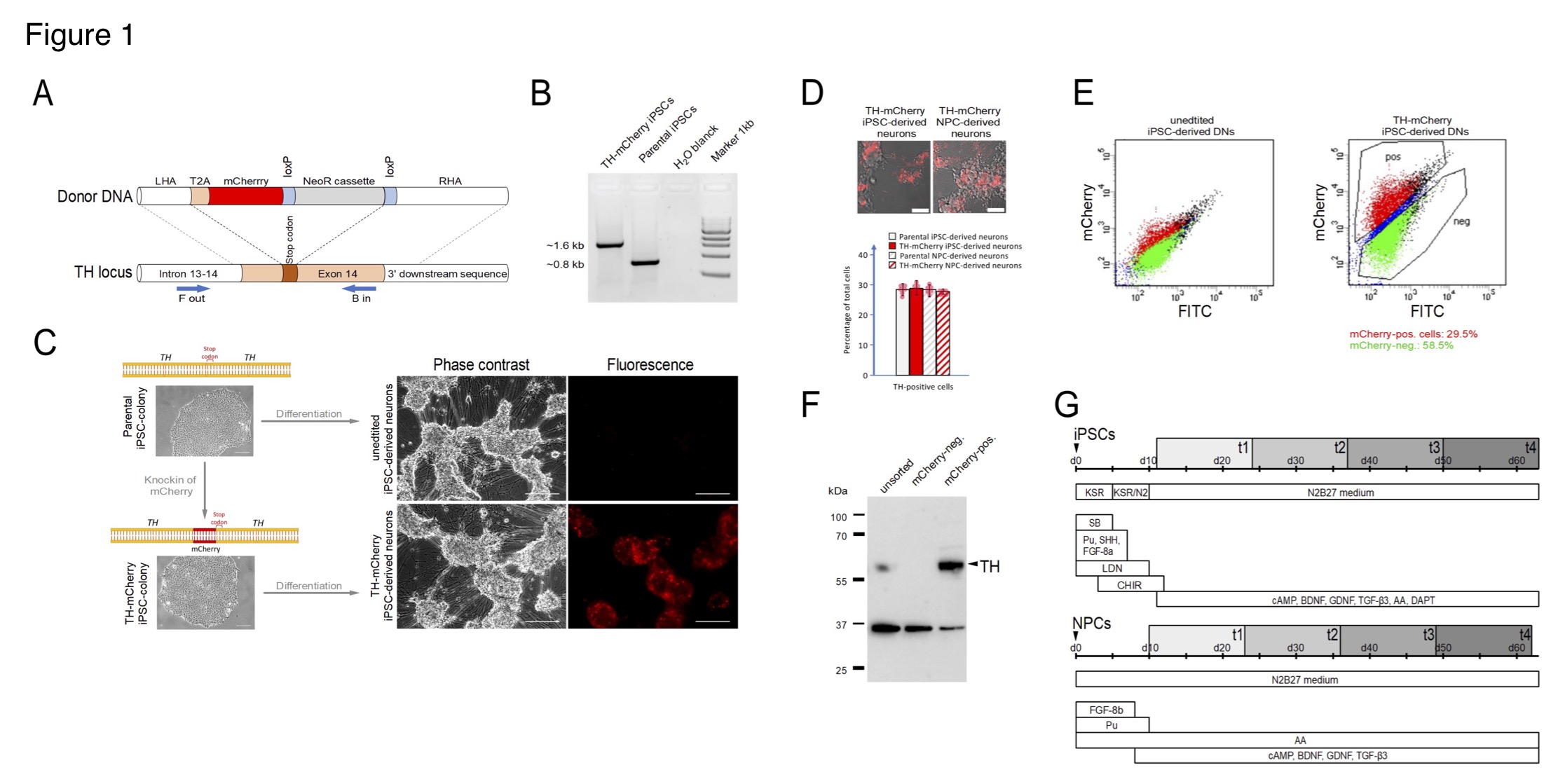

(A) Generation of TH-mCherry reporter iPSC and NPC lines and associated dopaminergic neuronal cultures. (A) Scheme illustrating the recombination strategy employed for genome editing. Blue arrows represent primers used for the PCR screening procedure. (B) PCR analysis of edited (TH-mCherry iPSCs) and unedited (parental iPSCs) iPSC lines confirming correct integration of the T2A-mCherry sequence in the TH locus of the parental iPSC line. (C) Phase contrast and associated fluorescence images of representative clusters of undifferentiated iPSC colonies (left) and terminally differentiated DN cultures (right), obtained from unedited (unedited iPSC-derived neurons) or edited (TH-mCherry iPSC-derived neurons) iPSCs (left). Scale bars: 100 µm. (D) Live-cell images of TH-mCherry reporter DN cultures derived from iPSCs or NPCs (top, scale bars: 50 µm) and analysis (bottom) of the percentage of TH-positive cells present in unedited and edited iPSC- and NPC-derived neuronal cultures, obtained from fixed samples immunostained with a TH-specific antibody. For each condition, a total of six replicates from two independent differentiations were evaluated. (E) Representative two-component density plots for FACS sorting of terminally differentiated parental (left) and TH-mCherry reporter iPSC cultures (right) using FITC- and mCherry-specific detection channels. The unspecific autofluorescence of unedited iPSC-derived neurons served as template to set the sorting gate for mCherry-negative cells and to define the gate corresponding to mCherry-positive cells. (F) Western blot analysis to evaluate TH protein levels (60 kDa) in unsorted, FACS-sorted mCherry-negative and mCherry-positive cells. GAPDH (36 kDa) served as loading control. (G) Timelines summarizing the two differentiation strategies used for the creation of iPSC- (top) and NPC-derived DNs. Electrophysiological analysis of cells was performed during an eight-week period (grey rectangles), t1-t4 specify the four timing groups.
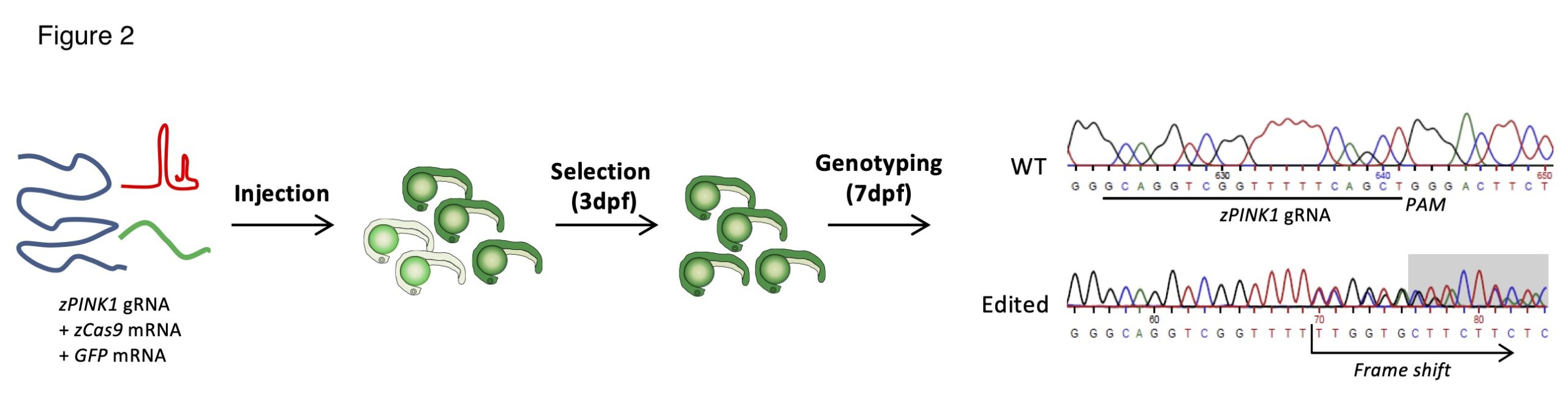

Generation of pink1 knockout Zebrafish lines.
- Translational Neurogenetics
- Genetics of Rare Diseases
- Functional Genetics of Movement Disorders
- Integrative Omics in Parkinson Disease
- Neuropsychiatric Epidemiology
- Clinical Neurogenetics and Neuroimaging
- Mitochondrial function in movement disorders
- Institut für Systemische Motorikforschung
- Applied Stem Cell Biology